Aptagen, LLC
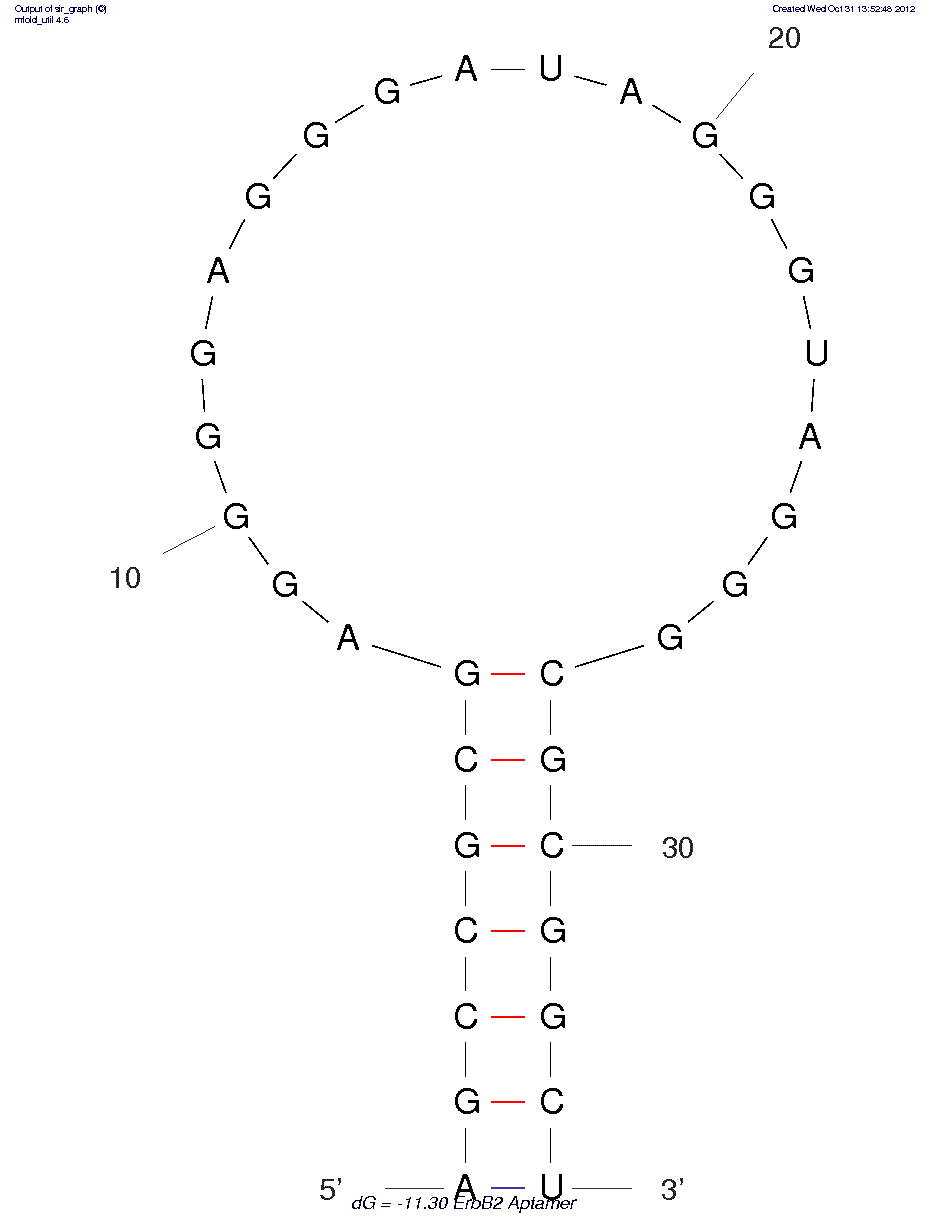 * The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
2'-F-RNA
ErbB2 in Breast Cancer Cells
Protein
3.49 nM (reported value)
Binding buffer: 20 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, and 40 U RNase Inhibitor
N/A°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
MDA-MB-453 breast cancer cell line, T47D breast cancer cells, and KPL-4 breast cancer cells (allErbB2 positive)showed staining, while ErbB2 negative cell line MCF-7 did not.
* The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
2'-F-RNA
ErbB2 in Breast Cancer Cells
Protein
3.49 nM (reported value)
Binding buffer: 20 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM MgCl2, 2 mM DTT, and 40 U RNase Inhibitor
N/A°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
MDA-MB-453 breast cancer cell line, T47D breast cancer cells, and KPL-4 breast cancer cells (allErbB2 positive)showed staining, while ErbB2 negative cell line MCF-7 did not.
ErbB2 (SE15-8) (ID# 202)

5'rAprGpfCpfCprGpfCprGprAprGprGprGprGprAprGprGprGprApfUprAprGprGprGpfUprAprGprGprGpfCprGpfCprGprGpfCpfUp3'
34
11239.85 g/mole
341100 L/(mole·cm)
73.53%
2.93
32.95
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Kim and Jeong. "In Vitro Selection of RNA Aptamer and Specific Targeting of ErbB2 in Breast Cancer Cells." Oligonucleotides, doi:10.1089/oli.2011.028.
Have your aptamer oligo synthesized ORDER NOW




We are always looking for ways to improve. Please tell us what you think.