Aptagen, LLC
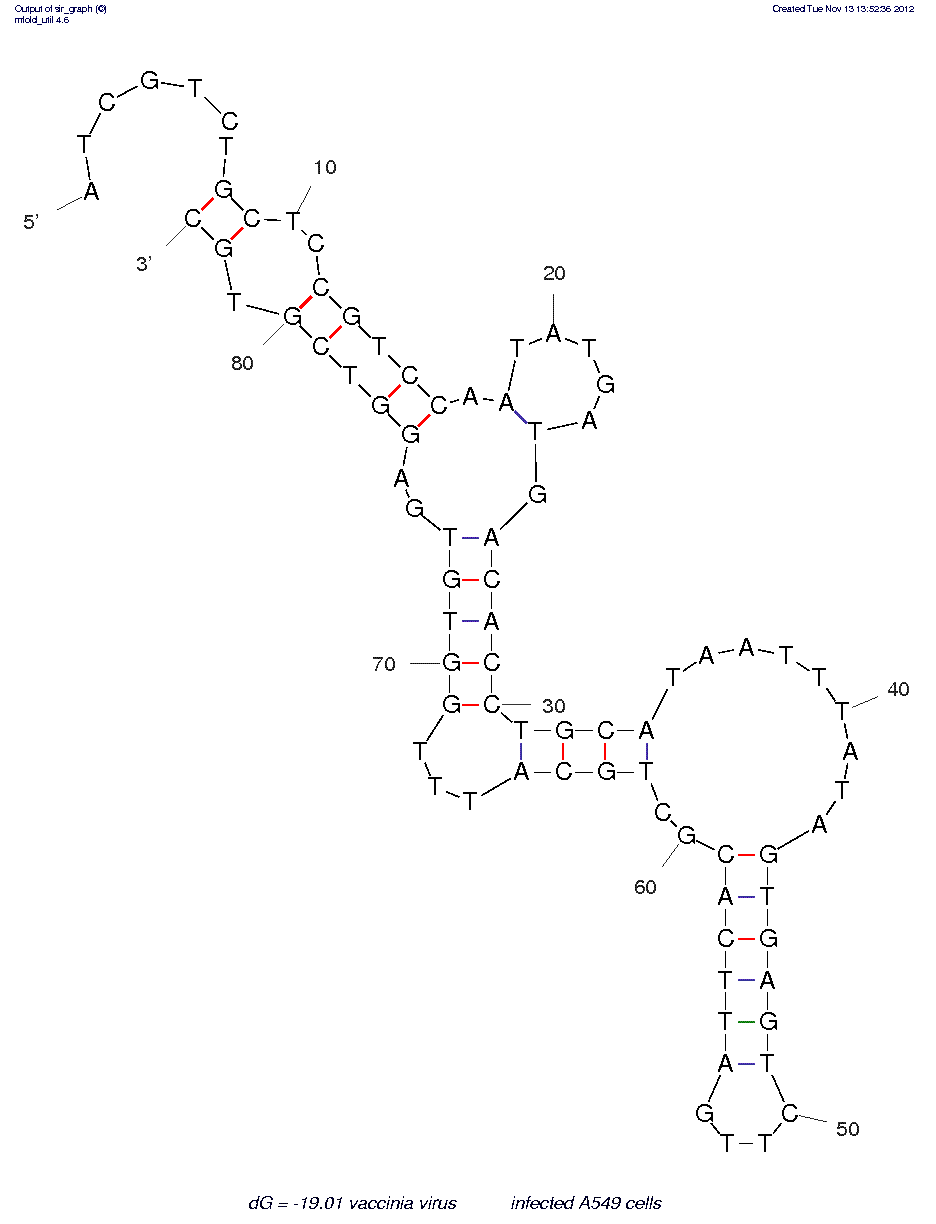
Vaccinia virusƒ??infected A549 cells (TV08) (ID# 7663)

DNA
Vaccinia virus (WR Strain)ƒ??infected A549 cells
Cells
2.7 nM (reported value)
Binding Buffer: Dulbecco PBS, 4.5g/L glucose, 5 mmol/L MgCl2, 0.1 g/L yeast tRNA, 100 mL/L FBS, and 1 g/L BSA.
4°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
The aptamer was also able to discriminate between infected and uninfected Hep 3B, Hep G2, HeLa, HCT 116, and CaOV3 cell lines and vaccinia IHDJ-infected cells.
5'-dApdTpdCpdGpdTpdCpdTpdGpdCpdTpdCpdCpdGpdTpdCpdCpdApdApdTpdApdTpdGpdApdTpdGpdApdCpdApdCpdCpdTpdGpdCpdApdTpdApdApdTpdTpdTpdApdTpdApdGpdTpdGpdApdGpdTpdCpdTpdTpdGpdApdTpdTpdCpdApdCpdGpdCpdTpdGpdCpdApdTpdTpdTpdGpdGpdTpdGpdTpdGpdApdGpdGpdTpdCpdGpdTpdGpdCp-3'


83
25569.65
784600
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Tang et al. "Generating Aptamers for Recognition of Virus-Infected Cells." Clin. Chem., 55(2009): 813-822.
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.