Aptagen, LLC
DNAzymeMB (Part of Lysozyme Aptasensor) (ID# 7581)

DNA
N/A: See additional comments.
Protein
0.1 fM (reported value)
NEBuffer 2(10X): 50 mM NaCl, 10 mM Tris-HCL, 10 mM MgCl2 1 mM dithiothreitol, pH 7.9.
Detection Buffer (total volume): 25 mM HEPES, 20 mM KCL, 200 mM NaCl, 0.05% Triton X-100 and 1% DMSO; pH 7.4) with 0.5 µM hemin.
Consult ref. for procedure.
Detection Buffer (total volume): 25 mM HEPES, 20 mM KCL, 200 mM NaCl, 0.05% Triton X-100 and 1% DMSO; pH 7.4) with 0.5 µM hemin.
Consult ref. for procedure.
RT°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
BSA was used as a negative control.
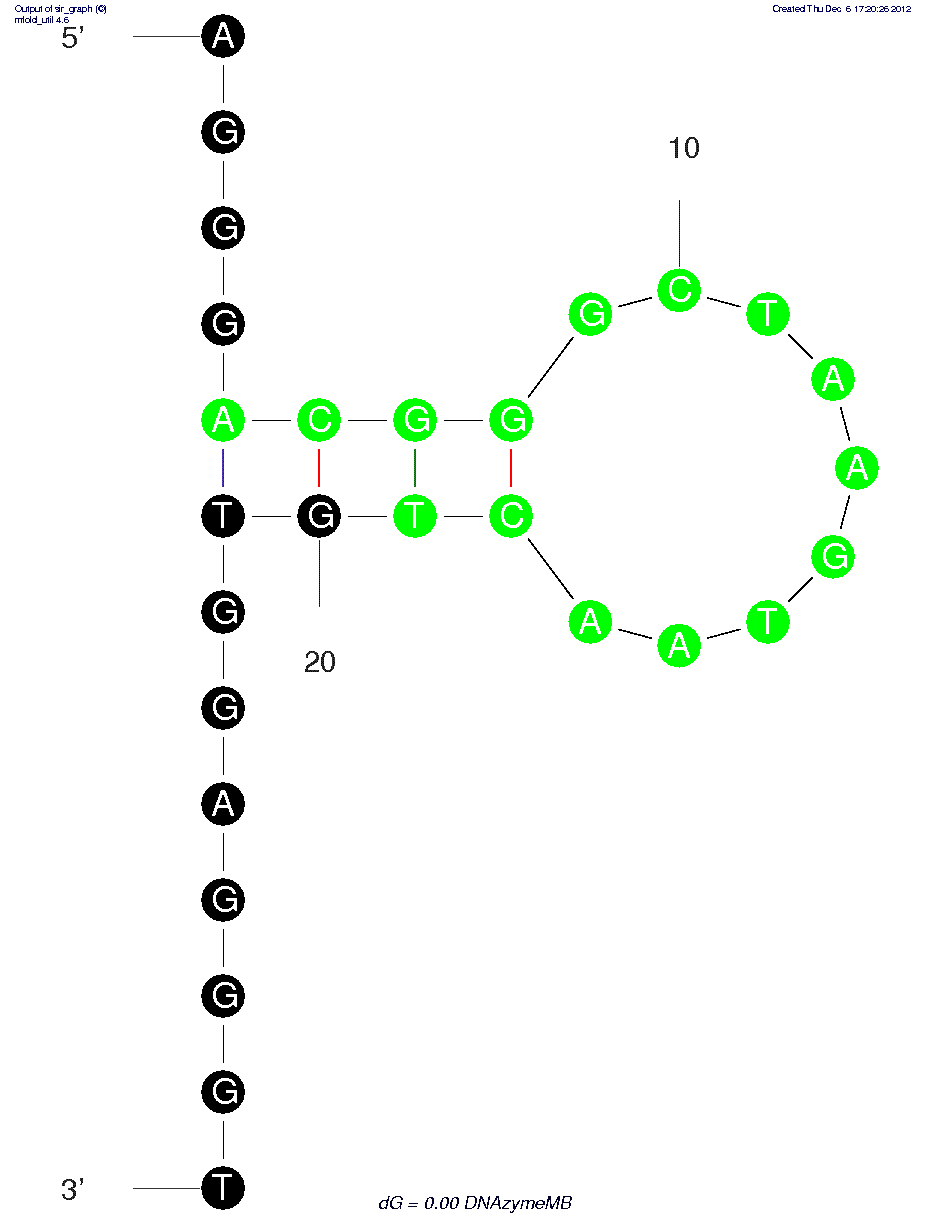
Contains a horseradish peroxidase-like DNAzyme with a region complementary to the Lysozyme Aptamer DNA Blocker (highlighted green in the structure). In the presence of the blocker, the two oligonucleotides hybridize, inactivating the DNAzyme. When the target lysozyme is added, DNAzyme-Blocker complex dissociates and the DNAzyme is activated. For more details, consult ref.
5'-dApdGpdGpdGpdApdCpdGpdGpdGpdCpdTpdApdApdGpdTpdApdApdCpdTpdGpdTpdGpdGpdApdGpdGpdGpdTp-3'


28
8798.78
285700
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Fu et al. "An ultrasensitive peroxidase DNAzyme-associated aptasensor that utilizes a target-triggered enzymatic signal amplification strategy." Chemical Communications, 47(2011): 9876-9878.
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.