Aptagen, LLC
CSS.1 (ID# 8201)

DNA
Cortisol
Small Organic
245 nM (reported value)
To study aptamer binding under different conditions, a number of buffers were prepared. Buffer 1 (the buffer used for selecting the CSS.1 aptamer): 20 mM HEPES (pH 7.5), 5 mM KCl, 1 M NaCl and 10 mM MgCl2. Buffer 1-1: 20 mM HEPES (pH 7.5), 5 mM KCl, 100 mM NaCl and 10 mM MgCl2. Buffer 1-2: 20 mM HEPES (pH 7.5), 5 mM KCl, 100 mM NaCl and 2 mM MgCl2. Buffer 1-3: 20 mM HEPES (pH 7.5), 5 mM KCl and 100 mM NaCl. Buffer 2 (the buffer used for selecting the 15-1 aptamer): 50 mM HEPES (pH 7.5), 137 mM NaCl and 5 mM MgCl2. Buffer 3 (the buffer used for avoiding the adsorption of HEPES on AuNPs): 10 mM phosphate (pH 7.5) and 1 mM MgCl2. Artificial interstitial fluids (ISF): 2.5 mM CaCl2, 10 mM HEPES, 3.5 mM KCl, 0.7 mM MgSO4, 123 mM NaCl, 1.5 mM NaH2PO4, 7.4 mM sucrose, pH 7.5.38
25°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
N/A
Na+ and Mg2+ were not required for CSS.1, but Mg2+ was helpful to ensure a high binding affinity.
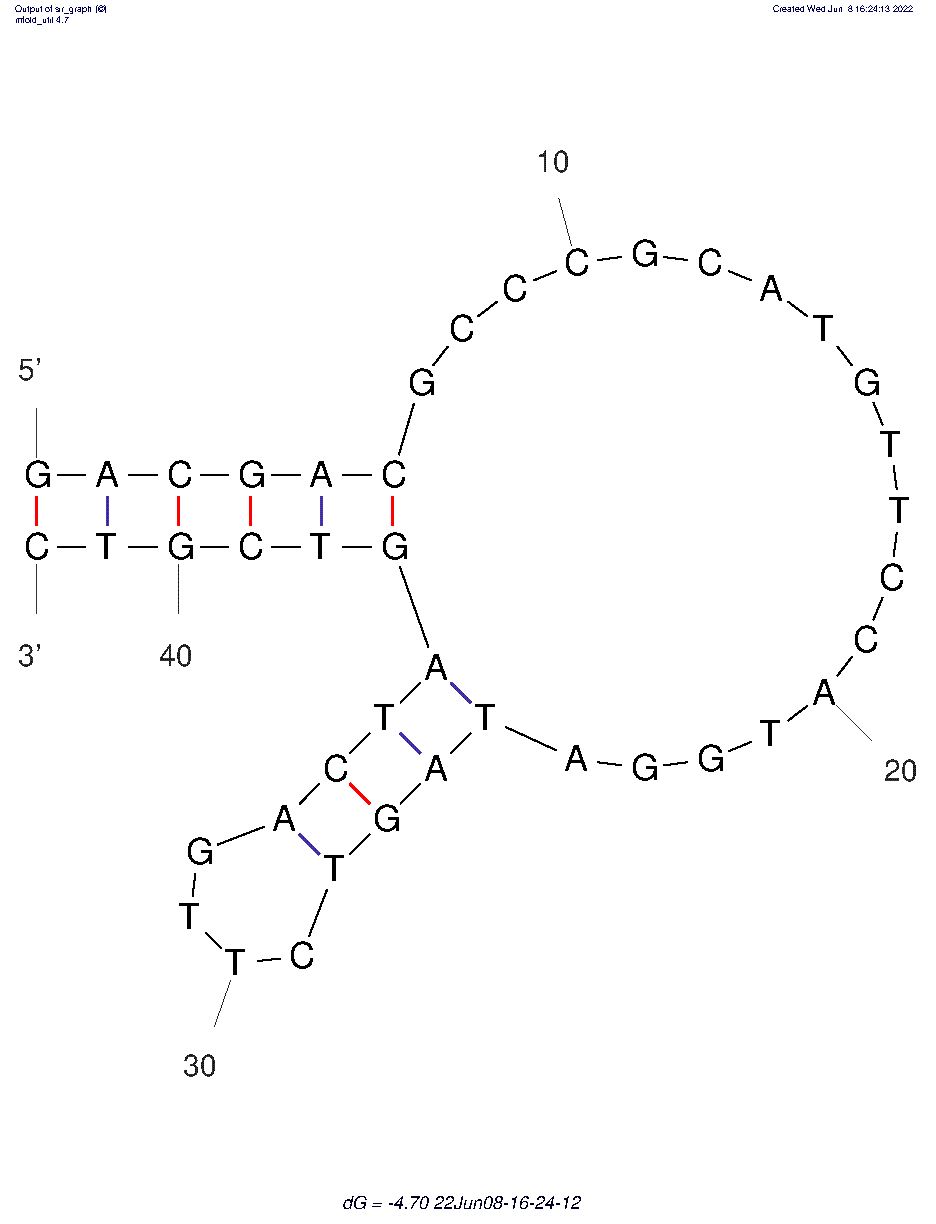
5'-dGpdApdCpdGpdApdCpdGpdCpdCpdCpdGpdCpdApdTpdGpdTpdTpdCpdCpdApdTpdGpdGpdApdTpdApdGpdTpdCpdTpdTpdGpdApdCpdTpdApdGpdTpdCpdGpdTpdCp-3'


42
12881.4
395100
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Niu, C., Ding, Y., Zhang, C., & Liu, J. (2022). Comparing two cortisol aptamers for label-free fluorescent and colorimetric biosensors. Sensors & Diagnostics, 1(3), 541–549. https://doi.org/10.1039/d2sd00042c
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.