Aptagen, LLC
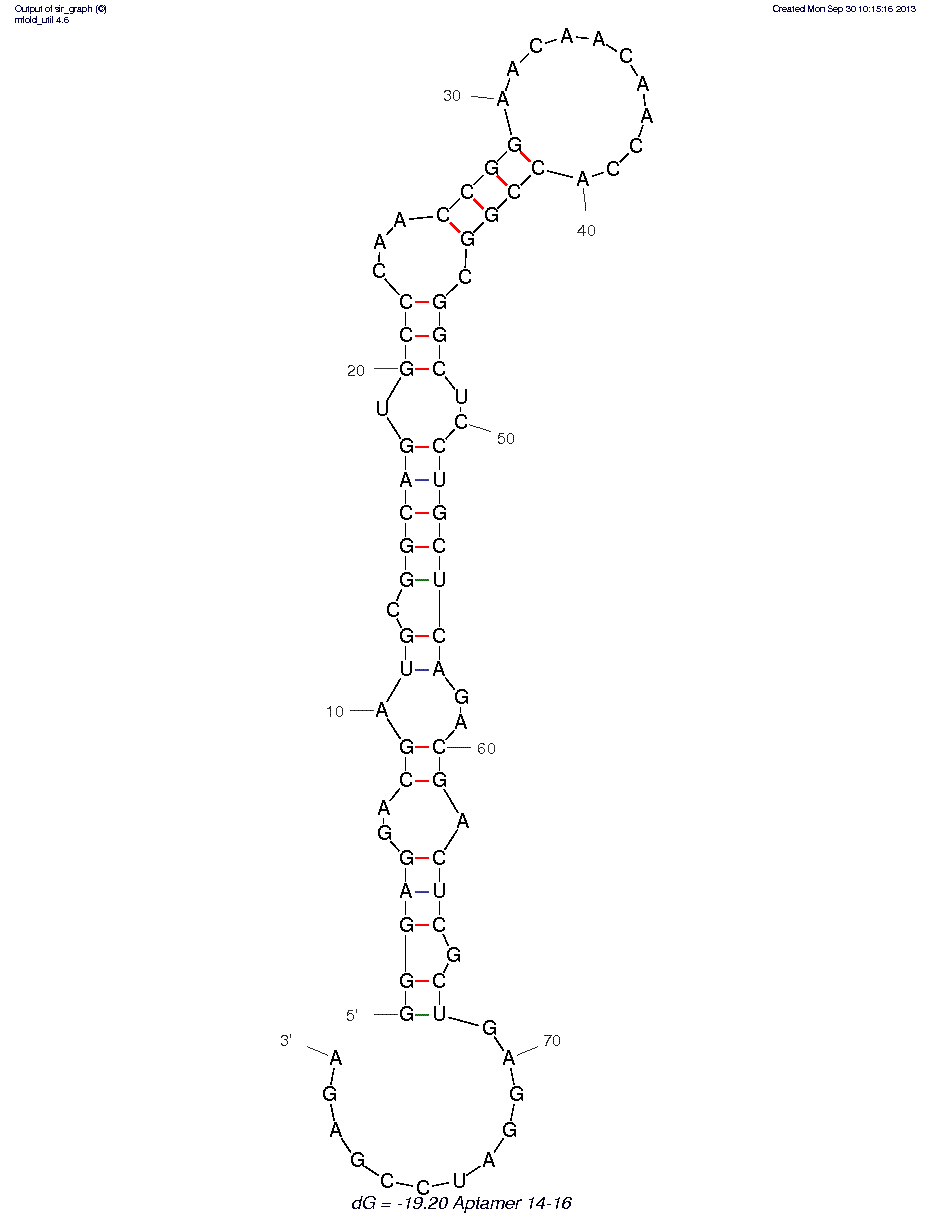 * The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
* The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
14-16 (ID# 7865)

2'-F-RNA
Mouse P68 RNA Helicase (Ddx5)
Protein
30.8 nM (reported value)
For selection: 5 nmol of aptamer pool in 200 µl of PBS, injected systemically and allowed to circulate for 20 minutes For binding assay: 10 mM Tris-HCl (pH 7.0), 150 mM NaCl, 10% (v/v) glycerol, 2.5 µg/ml tRNA. 10 minute incubation at 37°C For affinity purification: 20 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM CaCl2, 0.1% BSA, 0.1 µM tRNA at 4°C for 2 hours
37°C
N/A
See additional comments. Initially demonstrated twofold increased affinity (30.8 nM) for aggregate CT26 tumor proteins over normal colon tissue (68.9 nM).
Has better affinity for HA-tagged version of human p68 protein (13.8 nM), but only has an affinity of 190 nM for HA-tagged Tenascin CD. ATPase activity was significantly decreased in the presence of aptamer 14-16. IC50 of aptamer was 0.78 µM compared to that of the library (3.63 µM). An additional ƒ??Uƒ? nucleotide is present at position 55 in supplementary Figure 1-b and in the later-published version of the paper available at http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2795795/. This sequence was used to predict the secondary structure.
5'-rGprGprGp rAprGprGp rApfCprGp rApfUprGp fCprGprGp fCprAprGp fUprGpfCp fCpfCprAp rApfCpfCp rGprGprAp rApfCprAp rApfCprAp rApfCpfCp rApfCpfCp rGprGpfCp rGprGpfCp fUpfCpfCp fUprGpfCp fUpfCprAp rGprApfCp rGprApfCp fUpfCprGp fCpfUprGp rAprGprGp rApfUpfCp fCprGprAp rGprAp-3'


80
25949.66
769500
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Mi et al. ƒ??In vivo selection of tumor-targeting RNA motifsƒ?. Nature Chemical Biology 1(2010): 22-24. DOI: 10.1038/NCHEMBIO.277
Have your aptamer oligo synthesized ORDER



We are always looking for ways to improve. Please tell us what you think.