Aptagen, LLC
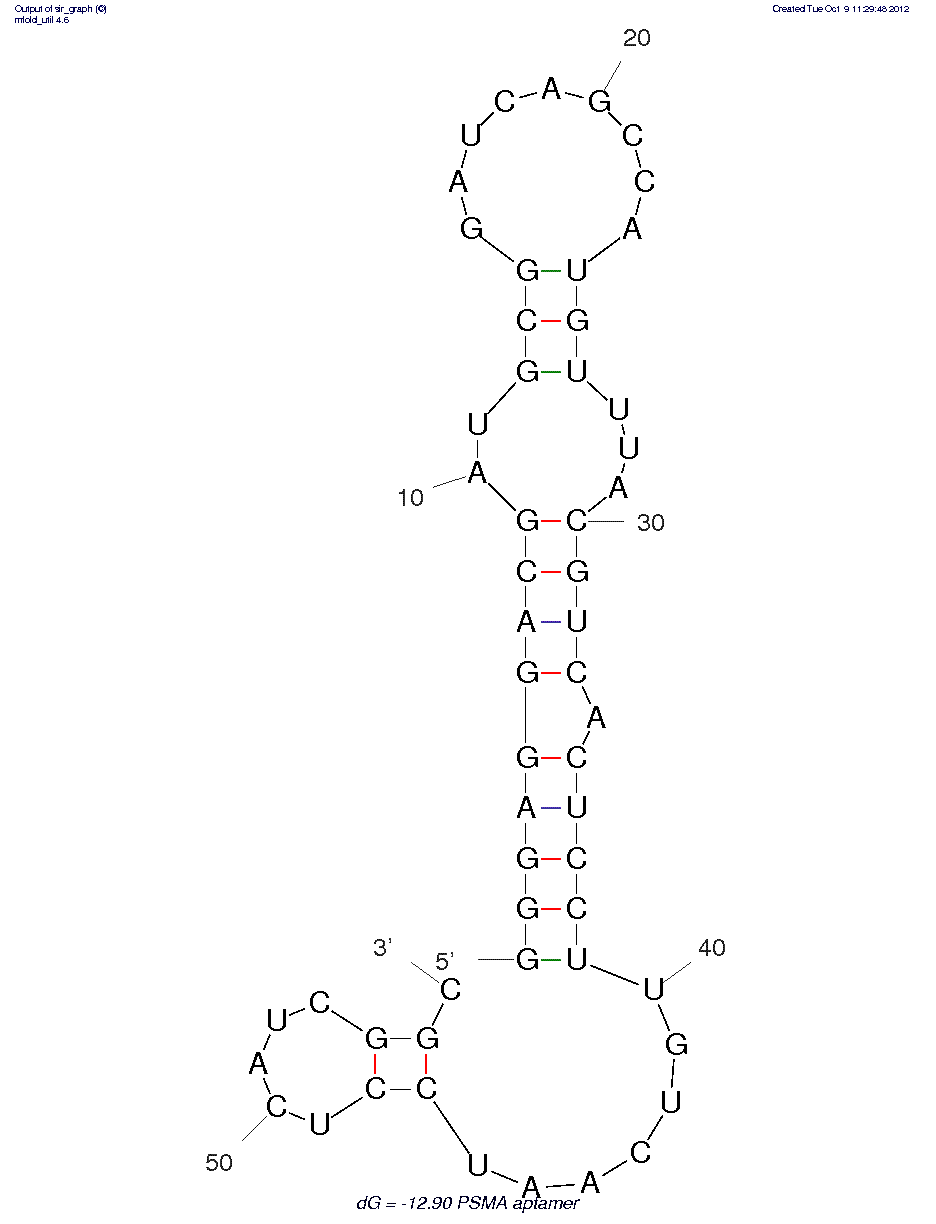 * The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
2'-F-RNA
PSMA
Protein
20.5 nM (reported value)
HEPES buffered saline (pH 7.4), 1 mM MgCl2, 1 mM CaCl2, 0.01% I-block (Tropix Inc. Bedford, MA), and 0.05% Tween 20. Incubated at 37 °C for 30 minutes. Washed using the above buffer at 37 °C.
N/A°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
Truncated aptamer is given. A higher Ki was found for the full-length aptamer; see reference for details.
* The secondary structure may not be accurate because of MFOLD limitations on chemistries other than RNA/DNA.
2'-F-RNA
PSMA
Protein
20.5 nM (reported value)
HEPES buffered saline (pH 7.4), 1 mM MgCl2, 1 mM CaCl2, 0.01% I-block (Tropix Inc. Bedford, MA), and 0.05% Tween 20. Incubated at 37 °C for 30 minutes. Washed using the above buffer at 37 °C.
N/A°C
NA If the oligo is a known aptamer sequence: For binding studies, perform a refolding protocol to ensure proper function (i.e. binding to antigen or target). Refer to the aptamer reference source for the appropriate refolding parameters and binding conditions. Note: it is unknown whether aptamer functions properly without refolding.
Truncated aptamer is given. A higher Ki was found for the full-length aptamer; see reference for details.
PSMA Aptamer (A10-3) (ID# 83)

5'rGprGprGprAprGprGprApfCprGprApfUprGpfCprGprGprApfUpfCprAprGpfCpfCprApfUprGpfUpfUpfUprApfCprGpfUpfCprApfCpfUpfCpfCpfUpfUprGpfUpfCprAprApfUpfCpfCpfUpfCprApfUpfCprGprGpfCp3'
56
17966.62 g/mole
539200 L/(mole·cm)
55.36%
1.85
33.32
Note: Information on this aptamer oligo was obtained from the literature and hasn't been validated by Aptagen.
Lupold, Shawn, et al. "Identification and Characterization of Nuclease-stabilized RNA Molecules That Bind Human Prostate Cancer Cells via the Prostate-specific Membrane Antigen." American Association for Cancer Research Journal 62, (2002): 4029-4033
Have your aptamer oligo synthesized ORDER NOW




We are always looking for ways to improve. Please tell us what you think.